Professor, biology department, biology building room 313, extension 3887, cs10@aub.edu.lb
Research Interests
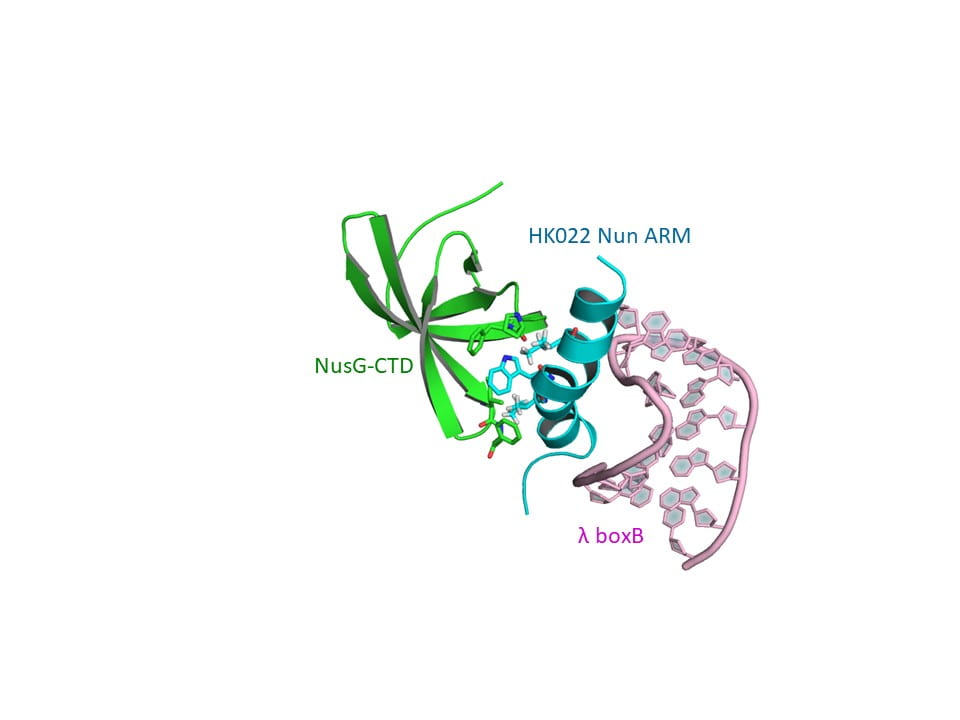
My broad interests lie in biochemistry and molecular evolution. Current projects study how viral RNA-protein interactions evolve new recognition strategies. Neutral theories of evolution contend that for any given genotype with a specific phenotype, there are sufficient single-site mutants of neutral fitness such that paths exist between genotypes with different phenotypes. Thus, by traversing intersecting paths, random change can lead to new phenotypes. Whether the validity of neutral theories extends beyond the predictable phenotypes of RNA secondary structure to more complex phenotypes typically found in viral regulation is not obvious. Complexes of arginine-rich peptides and small RNAs are attractive models to study recognition because they are structurally diverse, are found mediating important regulatory events, and alter conformations upon binding. A two-plasmid lambda phage N-nut antitermination reporter system with homologous and heterologous viral peptide-RNA interactions has been applied as an excellent tool to explore how recognition strategies may evolve. Other research interests include applications of molecular biology to natural history and steroid biotransformation.
Biographical Sketch
B.A. 1986, Chemistry, Reed College, Portland, Oregon;
Ph.D. 1993, Chemistry, Washington University, Saint Louis, Missouri;
Post-Doctoral Fellow, Department of Biochemistry and Biophysics, University of California, San Francisco;
Professor of Biology, American University of Beirut
Publications (Colin Andrew Smith at Google Scholar)
43. Raad NG, Ghattas IR, Amano R, Watanabe N, Sakamoto T, Smith CA.* 2020. Altered‐specificity mutants of the HIV Rev arginine‐rich motif‐RRE IIB interaction. Journal of Molecular Recognition, 33:e2833. doi: 10.1002/jmr.2833.
42. Baydoun E*, Wahab A*, Iqbal S, Smith C, Choudary MI. 2017. Biotransformation of drospirenone, a contraceptive drug, with Cunninghamella elegans. Steroids, 126, 30–34. doi: 10.1016/j.steroids.2017.07.010.
41. Haddad SG, Smith CA, Al Zein MS, Knio KM.* 2017. Genetic and morphometric variations in the Lebanese populations of the flower head-infesting tephritid, Terellia serratulae. Canadian Entomologist, 149, 73–88. doi:10.4039/tce.2016.26.
40. Baydoun E*, Wahab AT, Shoaib N, Ahmad MS, Abdel-Massih R, Smith C, Naveed N, Choudhary MI.* 2016. Microbial transformation of contraceptive drug etonogestrel into new metabolites with Cunninghamella blakesleeana and Cunninghamella echinulata. Steroids, 115, 56–61. doi: 10.1016/j.steroids.2016.08.003.
39. Itani GN, Smith CA.* 2016. Dust Rains Deliver Diverse Assemblages of Microorganisms to the Eastern Mediterranean. Scientific Reports 6, 22657. doi: 10.1038/srep22657.
38. Baydoun E*, Wahab A*, Mehmood H, Ahmad MS, Malik R, Smith C, Choudhary MI.* 2016. Microbial Transformation of danazol with Cunninghamella blakesleeana and anti-cancer activity of transformed products. Steroids, 105, 121–127. doi: 10.1016/j.steroids.2015.11.010.
37. Tawk CS, Ghattas IR, Smith CA.* 2015. HK022 Nun Requires Arginine-Rich Motif Residues Distinct from λ N. Journal of Bacteriology 197, 3573–3582, doi: 10.1128/JB.00466-15.
36. Baydoun E, Choudhary M, Wahab A, Smith C, Karam M, Farran D, Khan MSA, Ahmad MS. 2015. Treatment and inhibition of protozoal diseases with nandrolone and its derivatives. United States Patent and Trademark Office, US Patent 9,173,888.
35. Bariche M, Torres M, Smith C, Sayar N, Azzurro E, Baker R, Bernardi G.* 2015. Red Sea fishes in the Mediterranean Sea: a preliminary investigation of a biological invasion with DNA barcoding. Journal of Biogeography 42, 2363–2373. doi: 10.1111/jbi.12595.
34. Smith C, Wahab A*, Khan MS, Ahmad MS, Farran D, Choudhary MI, Baydoun E.* 2015. Microbial transformation of oxandrolone with Macrophomina phaseolina and Cunninghamella blakesleeana. Steroids 102, 39–45. doi: 10.1016/j.steroids.2015.06.008.
33. Abdallah EY, Smith CA.* 2015. Diverse Mutants of HIV RRE IIB Recognize Wild-Type Rev ARM or Rev ARM R35G-N40V. Journal of Molecular Recognition 28, 710‒721. doi: 10.1002/jmr.2485.
32. Baydoun E, Karam M, Atia-tul-Wahab A, Khan MS, Ahmad MS, Samreen, Smith C, Abdel-Massih R, Choudhary MI.* 2014. Microbial transformation of nandrolone with Cunninghamella echinulata and Cunninghamella blakesleeana and evaluation of leishmaniacidal activity of transformed products. Steroids 88, 95–100. doi: 10.1016/j.steroids.2014.06.020.
31. Yakoub S, El-Chami N, Kaszas K, Malek M, El Sirkasi M, Smith CA, Baydoun E, Tabone E, Manié SN, Régnier DCL.* 2014. The Proto-Oncoprotein c-Cbl Protects Cells against Oxidative Stress by Down-Regulating Apoptosis and is Highly Expressed in Several Cancers. Journal of Cancer Science & Therapy 6, 122–135. doi:10.4172/1948-5956.1000260.
30. Baydoun E, Bano S, Wahab AT, Jabeen A, Yousuf S, Mesaik A, Smith C, Choudhary I.* 2014. Fungal transformation and T-cell proliferation inhibitory activity of melengestrol acetate and its metabolite. Steroids 86, 56–61. doi: 10.1016/j.steroids.2014.04.012.
29. Possik EJ, Bou Sleiman MS, Ghattas IR, Smith CA.* 2013. Randomized Codon Mutagenesis Reveals that the HIV Rev Arginine-Rich Motif Is Robust to Substitutions and that Double Substitution of Two Critical Residues Alters Specificity. Journal of Molecular Recognition 26, 286–296. doi: 10.1002/jmr.2272.
28. Baydoun E, Bibi M, Iqbal MA, Wahab AT, Farran D, Smith C, Sattar SA, Rahman AU, Choudhary MI, Rahman A, Choudhary I.* 2013. Microbial transformation of anti-cancer steroid exemestane and cytotoxicity of its metabolites against cancer cell lines. Chemistry Central Journal 7:57. doi: 10.1186/1752-153X-7-57.
27. Bassil NM, Abdel-Massih R, El Chami N, Smith CA, Baydoun E.* 2012. Pleurotus ostreatus and Ruscus aculeatus Extracts Cause Non-Apoptotic Jurkat Cell Death. Journal of Plant Studies 1, 14–24. doi: 10.5539/jps.v1n1p14.
26. Haykal J, Geara F, Haddadin M, Smith CA, Gali-Muhtasib H.* 2009. The radiosensitizer 2-benzoyl-3-phenyl-6,7-dichloroquinoxaline 1,4-dioxide induces DNA damage in EMT-6 mammary carcinoma cells. Radiation Oncology 4, 25. doi: 10.1186/1748-717X-4-25.
25. Sayar NP, Smith CA, White IM, Knio KM.* 2009. Terellia fuscicornis (Diptera: Tephritidae): Biological and morphological adaptation on artichoke and milk thistle. Journal of Natural History, 43, 1159–1181. doi: 10.1080/00222930902807742.
24. Smith CA, Al Zein MS, Sayar NP, Knio KM.* 2009. Host Races in Chaetostomella cylindrica (Diptera: Tephritidae): Genetic and Behavioural Evidence. Bulletin of Entomological Research, 99, 425–32. doi: 10.1017/S0007485308006482.
23. Cocozaki AI, Ghattas IR, Smith CA.* 2008. The RNA-Binding Domain of P22 N Protein Is Highly Mutable and a Single Mutation Relaxes Specificity Toward λ. Journal of Bacteriology 190, 7699–7708. doi: 10.1128/JB.00997-08.
22. Asraoui J, Sayar NP, Khouzama KM, Smith CA.* 2008. Fly Diversity Revealed by PCR-RFLP of Mitochondrial DNA. Biochemistry and Molecular Biology Education 36, 354–362. doi: 10.1002/bmb.20220.
21. Cocozaki AI, Ghattas IR, Smith CA.* 2008. Bacteriophage P22 Antitermination BoxB Sequence Requirements Are Complex and Overlap with Those of λ. Journal of Bacteriology 190, 4263–4271. doi: 10.1128/JB.00059-08.
20. Haykal J, Fernainya P, Itani W, Haddadin M, Geara F, Smith C, Gali-Muhtasib H.* 2008. Radiosensitization of EMT6 Mammary Carcinoma Cells by 2-benzoyl-3-phenyl-6,7-dichloroquinoxaline 1,4-dioxide. Radiotherapy and Oncology 86, 412–418.
19. Smith C, Huppertz A, Wittig B. 2007. DNA Expression Construct for Multiple Gene Expression. European Patent Office, EP 1 530 641.
18. Schroff M, Smith C. 2006. Circular Expression Construct for Gene Therapeutic Applications. European Patent Office, EP 1 631 672.
17. Moreno S, Lopez-Fuertes L, Vila-Coro AJ, Sack F, Smith CA, Konig SA, Wittig B, Schroff M, Juhls C, Junghans C, Timon M.* 2004. DNA Immunisation with Minimalistic Expression Constructs. Vaccine 22, 1709–1716.
16. Junghans C, Schroff M, Koenig-Merediz SA, Alfken J, Smith C, Sack F, Schirmbeck R, Wittig B.* 2001. Form Follows Function: The Design of Minimalistic Immunogenically Defined Gene Expression (MIDGE®) Constructs. Plasmids for Therapy and Vaccination, edited by M. Schleef, Wiley-VCH Verlag GmbH.
15. Smith CA, Calabro V, Frankel AD.* 2000. An RNA-Binding Chameleon. Molecular Cell 6, 1067–1076.
14. Smith CA, Chen L, Frankel AD.* 2000. Using Peptides as Models of RNA-Protein Interactions. Methods in Enzymology 318, 423–438.
13. Peled-Zehavi, H, Smith CA, Harada K, Frankel AD.* 2000. Screening RNA-Binding Libraries by Transcriptional Antitermination in Bacteria. Methods in Enzymology 318, 297–308.
12. Smith CA, Crotty S, Harada Y, Frankel AD.* 1998. Altering the Context of an RNA Bulge Switches the Binding Specificities of Two Viral Tat Proteins. Biochemistry 37, 10808–10814.
11. Frankel AD,* Smith CA. 1998. Induced Folding in RNA-Protein Recognition: More than a Simple Molecular Handshake. Cell 92, 149–151.
10. Smith CA, Baeten J, Taylor JS.* 1998. The Ability of a Variety of Polymerases to Synthesize Past Site-Specific Cis-Syn, Trans-Syn-II, (6-4), and Dewar Photoproducts of TpT. Journal of Biological Chemistry 273, 21933–21940.
9. Jen J, Mitchell DL, Cunningham RP, Smith CA, Taylor JS, Cleaver JE.* 1997. Ultraviolet Irradiation Produces Novel Endonuclease III-Sensitive Cytosine Photoproducts at Dipyrimidine sites. Photochemistry and Photobiology 65, 323–329.
8. Smith CA, Wang M, Jiang N, Che L, Zhao X, Taylor JS.* 1996. Mutation Spectra of M13 Vectors Containing Site-Specific Cis-Syn, Trans-Syn-I, (6-4), and Dewar Pyrimidone Photoproducts of Thymidylyl-(3′-5′)-Thymidine in Escherichia coli under SOS Conditions. Biochemistry 35, 4146–4154.
7. Bowman KK, Sidik K, Smith CA, Taylor JS, Doetsch PW, Freyer GA.* 1994. A New ATP-Independent DNA Endonuclease from Schizosaccharomyces pombe that Recognizes Cyclobutane Pyrimidine Dimers and 6-4 Photoproducts. Nucleic Acids Research 22, 3026–3032.
6. Kim ST, Malhotra K, Smith CA, Taylor JS, Sancar A.* 1994. Characterization of (6-4) Photoproduct DNA Photolyase. Journal of Biological Chemistry 269, 8535–8540.
5. Reardon JT, Nichols AF, Keeney S, Smith CA, Taylor JS, Linn S, Sancar A.* 1993. Comparative Analysis of Binding of Human Damaged DNA-Binding Protein (XPE) and Escherichia coli Damage Recognition Protein (UvrA) to the Major Ultraviolet Photoproducts: T[c,s]T, T[t,s]T, T[6-4]T, and T[Dewar]T. Journal of Biological Chemistry 268, 21301–21308.
4. Kim ST, Malhorta K, Smith CA, Taylor JS, Sancar A.* 1993. DNA Photolyase Repairs the Trans-Syn Cyclobutane Thymine Dimer. Biochemistry 32, 7065–7068.
3. Smith CA, Taylor JS.* 1993. Preparation and Characterization of a Set of Deoxyoligonucleotide 49-mers Containing Site-Specific Cis-Syn, Trans-Syn-I, (6-4), and Dewar Photoproducts of Thymidylyl(3′-5′)-Thymidine. Journal of Biological Chemistry 268, 11143–11151.
2. Svoboda DL, Smith CA, Taylor JS, Sancar A.* 1993. Effect of Sequence, Adduct Type, and Opposing Lesions on the Binding and Repair of Ultraviolet Photodamage by DNA Photolyase and (A)BC Excinuclease. Journal of Biological Chemistry 268, 10964–10700.
1. Bennet RM,* Hefeneider SH, Bakke A, Merrit M, Smith CA, Mourich D, Heinrich MC. 1988. The Production and Characterization of Murine Monoclonal Antibodies to a DNA Receptor on Human Leukocytes. Journal of Immunology 140, 2937–2942.